Exploring microbial diversity in South Korean caves through shotgun sequencing: contrasting dry and wet environments, swabbing versus sediment sampling
Copyright ⓒ The Geological Society of Korea 2024
Abstract
Caves host bacterial communities that represent diverse and unique ecosystems, often isolated from external influences. Diversity among bacterial colonies can vary greatly depending on the cave’s geographical location but it is also linked to the environmental conditions inside the cave such as a depth gradient, light and humidity levels, nutrients availability and substrate conditions. Microbes in caves have adapted to live under extreme conditions and because of that have often developed unique metabolic pathways. In this present work, metagenomics sequence data from five South Korean caves are analyzed with the main objective of identifying microbial taxa as potential proxies for cave environmental conditions. Two independent shotgun sequencing platforms have been used targeting short and long reads through Illumina and Oxford Nanopore technologies respectively. The DNA samples come from swabs of rock surfaces or sediments inside these Korean caves that can be classified as wet, dry or seasonally wet. The internal depth gradient as well as different substrates (swabs and sediments) have also been considered for the interpretation of the composition of the internal microbiota. Dry environments like those from Baram and Simbok Caves have a high abundance of species belonging to the phylum Actinobacteria, compared to wetter caves like Ondal, Sanjidang and Seodae where Proteobacteria are instead more abundant. In dark and undisturbed sectors of the caves, the microbial colonies are dominated by Gammaproteobacteria compared to areas near the entrance.
초록
동굴은 흔히 외부환경의 영향으로부터 격리되어 다양하면서도 독특한 생태계와 박테리아 군집을 이룬다. 박테리아 군체 사이의 다양성은 동굴의 지리적 위치에 따라 크게 좌우될 수 있으나 동굴 입구로부터의 깊이, 빛, 습도, 영양공급 정도 및 기저 환경등 동굴 내부에서의 세밀한 환경 조건에도 영향을 받게 된다. 이에 따라 동굴 미생물은 특수한 대사 경로를 발달시킴으로써 극한환경 조건에 적응해왔다. 이번 연구에서는 동굴환경 조건에 대한 잠재적 지시자로서의 미생물 분류군을 확인하기 위해 남한 지역의 5개 동굴로부터 채취한 시료들의 군유전체 염기서열을 분석하였다. 이를 위해 독립적 샷건 시퀀싱 플랫폼인 Illumina와 Oxford Nanopore technologies가 쇼트 및 롱 리드를 판독하는 데 적용되었다. 디옥시리보핵산 시료는 습윤, 건조, 계절적 습윤환경으로 각각 분류할 수 있는 동굴 내부 지점들로부터 암석표면 면봉긁기 방식과 퇴적물 채취 방식으로 확보하였다. 입구로부터의 깊이와 기저 환경(암석 또는 퇴적물) 조건을 중심으로 내부 미생물군 조성의 원인을 해석하였다. 연구 결과, 상대적으로 건조환경에 해당하는 바람굴 및 심복굴에서는 Actinobacteria 문에 속하는 종들이 우세하게 나타난 반면, 습윤환경으로 분류된 온달굴, 산지당굴, 서대굴에서는 Proteobacteria 문에 속하는 종들이 우세한 것으로 나타났다. 또한, 비교란 암흑대에서의 미생물 군집은 입구 인근 구역에 비해 Gammaproteobacteria 문이 우점하는 특징을 보인다.
Keywords:
cave metagenomics, South Korea, shotgun sequencing키워드:
동굴 군유전체학, 남한, 샷건 시퀀싱1. Introduction
Caves are considered oligotrophic environments due to their limited nutrient and energy sources to support life. Nevertheless bacteria, which constitute the major portion of cave biodiversity, can survive and thrive in this type of environment by modifying their metabolic strategies and microbial communities are also key organisms in the subterranean food web and in decompositions processes (Graening and Brown, 2003). In karstic environments, bacteria are affected by the geology of the surroundings and by the presence of specific geochemical elements and can utilize inorganic compounds for their energetic sustainment (Barton, 2006; De Mandal et al., 2017). Trophic levels and light intensity define the different zonal gradients within a cave and influence the distribution and abundance of the microbiota on the different substrates (Ghosh et al., 2017).
The cave's interior can be categorized into four primary zones based on light penetration: 1) the entrance zone; 2) the twilight zone, characterized by diminishing light until; 3) the transition zone, which remains dark but still experiences variations in temperature and moisture; and 4) the deep zone, where darkness is absolute, humidity is high, and the temperature remains constant (Ghosh et al., 2017).
Since microorganisms need to survive in low nutrient environments, they can use minerals and inorganic matter for catalyzing reactions (Dong et al., 2020) and are also involved in many in-cave mineralization processes. For example, calcifying bacteria promote carbonate precipitation (Cacchio et al., 2003), possibly in the form of speleothems (Cacchio et al., 2004), and/or catalyze the development of crystalline fabrics in stalagmites via metabolic processes or, intrinsically, via water evaporation and CO2 degassing (Franchi and Frisia, 2020).
On the other hands, heterotrophic microorganisms, including certain bacterial groups and fungi, thrive in caves on the abundant organic matter of substrates like guano, utilizing organic carbon as a source of energy (Dimkić et al., 2021). Since cave substrates are abundant in inorganic molecules such as iron (Parker et al., 2022), manganese (Carmichael et al., 2013), ammonia (Martin-Pozas et al., 2023), methane (Webster et al., 2022) and sulphur (Jones et al., 2016), chemolithoautotrophic organisms are particularly abundant in caves.
To overcome growth limiting factors, microorganisms establish sophisticated, mutualistic networks. This strategy enables a greater number of organisms to survive and grow in the extreme conditions of caves. On cave walls and speleothems, multispecies biofilms consisting of algae/ cyanobacteria, bacteria, or fungi, promoting nutrient circulation enabling diverse organisms to thrive in this harsh environment. Conversely, some bacteria favour competition over cooperation by producing secondary metabolites that suppress the growth of nearby microorganisms, particularly fungi (Roldán et al., 2009; Jaroszewicz et al., 2021; Kosznik-Kwaśnicka et al., 2021).
The main groups described in the cave ecosystem are Actinobacteria, Proteobacteria, Bacteriodetes, Acidobacteria, Firmicutes (Kato et al., 2024). Actinobacteria are able to capture CO2, which is highly abundant in the cave air and in turn promoting the precipitation of calcium carbonate and the emergence of speleothems (Cuezva et al., 2012).
Proteobacteria, the second largest phyla in caves, thrive in sediments, water, and rocks and can metabolize methane as their sole carbon and energy source (Zhu et al., 2021).
Moreover, heterotrophic bacteria in caves can be pathogenic and may contribute to emerging diseases, such as the Firmicutes phylum that thrive in substrates with high organic content, like close to waste disposal (Tomova et al., 2013) in caves frequently visited by people.
In this study we describe microbial communities from different South Korean caves and explore whether there is a correlation to different environmental conditions. Particularly, we wanted to identify possible differences in microbial composition between dry and wet caves and along the depth-gradient inside the caves. Two types of cave have been chosen: caves not accessible to the public and touristic show-caves, to inspect whether human disturbances impact the composition of bacterial communities.
Different sampling methods can yield varying insights into microbial communities; therefore two types of samples were selected, cave sediments and swabs of surfaces. Swabbing is often used to collect surface-associated microbes, providing information on microbial colonization patterns on cave walls and formations. In contrast, sediment sampling captures microbes residing within the cave substrates.
To have a better insight into the metagenomic reconstructions, shotgun metagenomic DNA sequencing based on two independent methods have been used: PCR-free long-reads sequencing with an Oxford Nanopore Technologies (ONT) MinION sequencer using sediment samples only and short-reads sequencing using an Illumina sequencing platform applied to only swabs. ONT MinION Technology major drawback is the high error rate compared to other sequencing technologies nevertheless, this has been consistently improved over the years with the release of new improved flow cells and more accurate basecalling software (Ciuffreda et al., 2021). At the current time of this work, the read accuracy is at 97%. The Illumina platforms, while performing better in terms of read accuracy and error rate (> 99%), when use short read lengths (150-300 bp) make it difficult to resolve complex genomic regions, such as those containing repetitive elements or large structural variants (Chiang and Dekker, 2020).
South Korean cave systems have not received much international scientific attention. International peer-reviewed studies on South Korean caves have so far focused on the microbial diversity found in moonmilk encrustations from Baeg-nyong Cave (Park et al., 2020), biota found in caves from the Gangwon and Chungcheongbuk provinces (Lee et al., 2020; Chang et al., 2021) and speleothem-based paleoclimate reconstructions by Jo et al. (2010, 2014, 2017). With this paper we aim to add to the knowledge about South Korea as a region of interest for cave systems based environmental reconstructions. Specifically, this work has been intended for paving the way to paleoenvironmental research using stalagmites as a source of ancient DNA (aDNA) and explore their potential as biological paleoarchives. Since the preservation of the aDNA is favored in deposits deeper inside caves (Stahlschmidt et al., 2019), it is crucial to understand the diversity of the local microbial communities and how the in-cave depth gradient plays a role. Some of the caves studied here showed a clearer microbial gradient compared to others but they all seem to agree that relatively drier surfaces are characterized by a preponderance of microbes belonging to the phylum Actinobacteria (also known as Actinomycetes) also confirmed by previous studies (Vardeh et al., 2018; Park et al., 2020).
2. MATERIALS AND METHODS
2.1. Geographical context and description of the caves
The present work examines the microbial communities of five South Korean caves from the central area of the country (Fig. 1) that have been divided into three categories: dry, seasonally wet, and permanently wet.
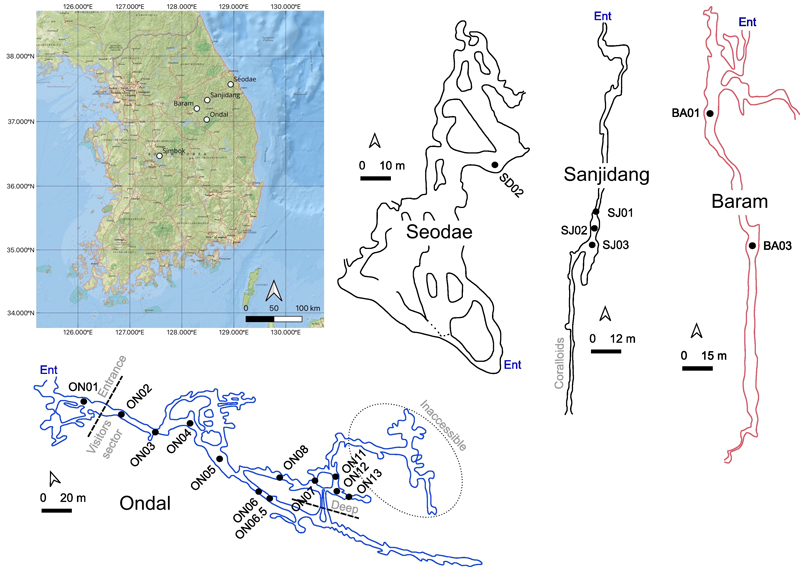
Cave maps with their geographical locations in the Korean peninsula (map from ESRI National Geographic, obtained using QGIS software). Blue outline for Ondal Cave (wet cave), red outline for Baram Cave (dry cave) and black outline for Seodae and Sanjidang Caves (seasonally wet caves). Simbok Cave map is unreported therefore is has been excluded from this figure. Maps have been modified from the originals. Surveyors: Kyoung-nam Jo and Moo-Yeol Lee (Seodae Cave map), Ki-Bok Kim and Sang-Ho Kim (Baram Cave map), Korea Cave Research Institute (Sanjidang and Ondal Cave maps).
Ondal Cave (ON) located in Danyang-gun, Chungcheongbuk-do (37°03.32'N, 128°29.27'E), is a show-cave open all year round to the public excluding during the monsoon season, when it can get flooded (Fig. 1). Like most Korean caves, Ondal does not extend to any considerable depth being developed only for less than 1 km. Air parameters monitoring has been performed in four occasions between November 2020 and April 2022 and showed a constant temperature of ~13℃ in the inner parts of the cave and high CO2 pressure, ranging from 460 ppmv up to 1100 ppmv due to seasonal ventilation processes. The accessible level of the cave corresponds to the phreatic zone with the subterranean river constantly flowing inside the cave and occasionally inundating parts of the cave. The cave is extremely well decorated with a wide range of deposits.
Simbok Cave (SB) (36°47.13'N, 127°57.76'E) is a small cave located in Goesan-gun, Chungcheongbuk-do in central South Korea (Lee et al., 2020). It is formed by horizontal passages accessible to humans in a standing position. During the time of the visiting and the sampling (October 2020), the cave was mostly dry. Hibernating bats were also observed hanging from the ceiling. For this cave no planimetry has been found but its length can be approximated to be nearly 500 meters.
Baram Cave (BA) (37°20.07'N, 128°29.13'E) (“wind cave” in Korean) is in the Gangwon province but further inland compared to Seodae Cave, in the Pyeongchang-gun county (Kim et al., 2018; Chang et al., 2021). The cave is formed by a single long and horizontal duct (~350 m long), which is quite wide and has high ceiling allowing an easy traversing. Towards the end of the cave, red-clay deposits pile up along the edges of the pavement and in the interstices. At the time of the visit, the cave was dry and excluding a few stalactites and broken stalagmites (only a couple of speleothems noticed) the cave is not decorated with any dripstones, and the bare bedrock is exposed. In some part of the cave, it is possible to observe some anticline fold structures and fault planes.
Seodae Cave (SD) is a cave in Gangwon province located in the southern part of the Seokbyeongsan Formation (37°57'N, 128°94'E) (Choi et al., 2003) (Fig. 1). The access to the cave is technical challenging and requires rope maneuvers. The structure of the system is quite complex with a combination of vertical and horizontal passages that extend for almost 800 meters. At the time of the exploration, the cave was dry, and no speleothem formations have been observed but during the summer monsoon season the drippings restore, suggesting a fast discharge rate of the vadose layer and seasonally modulated environmental conditions.
Sanjidang Cave (SJ) (37°49.04'N, 128°54.23'E) has multi-leveled, narrow and short (~ 520 meters) passages that mostly develop as a deep crack in the bedrock and “crevasses” obstruct the passing along the conduit (Fig. 1). The cave is peculiar because of coralloid deposits that entirely cover the walls in the deep sectors. At the time of the expedition, no air flows have been perceived by the participants, but the nature of coralloid speleothems indicates the occurrence of some sort of air flow or aerosols combined to active dripping (Vanghi et al., 2017), which most likely occurs during the wet season in summer.
2.2. Samples collection and DNA extraction
For each cave, around 20 grams of sediment were collected using a steel spatula decontaminated with bleach between different sampling actions and put into 50 ml polypropylene tubes. Sterile cotton tip swabs were used to sample areas of speleothem surfaces (5 cm2). To maximize the capture of the microbial diversity inside the cave, different speleothem deposits were swabbed, such as stalagmites, stalactites and flowstones. After swabbing the surface, each swab was placed into 2 ml vials filled with Zymo DNA/RNA shield buffer. The wooden sticks of each swab were then broken by hand making sure not to touch the cotton head submerged in the buffer, and the vials were finally closed with a screwtop. Once in the laboratory, the sediment samples were stored at -20℃ while the tubes with the swabs in DNA/RNA shield buffer were stored in the fridge at 5℃.
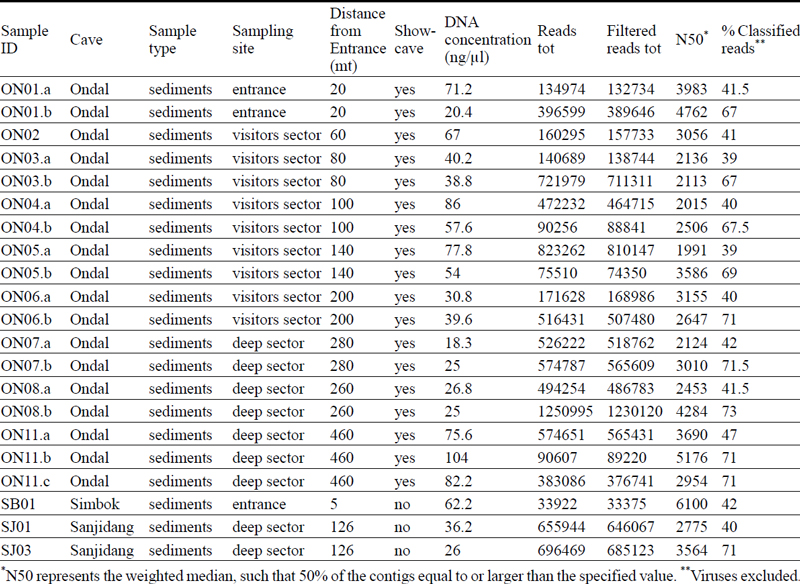
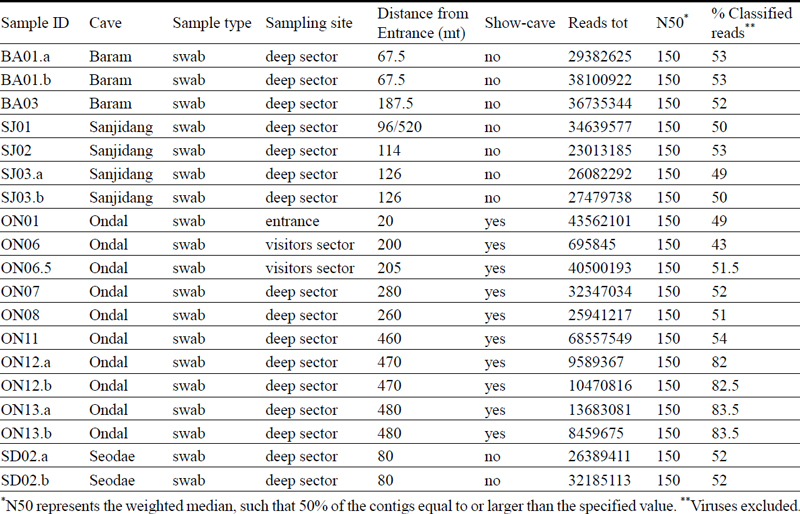
A total of 40 samples were collected at different depths inside the caves and named with two letters referring to the cave name and a number gradually increasing with increasing depth inside in the cave. For example, ON01 corresponds to a sample that was taken from a location inside Ondal Cave closer to the entrance compared to ON07 that was taken at a greater distance. Total number and labelling of the samples for each cave are indicated in Table 1 and 2.

Information about the samples (tot. 21) that have been sequenced using MinION sequencing technology.

Information about the samples (tot. 19) that have been sequenced using Illumina sequencing technology.
Some samples have biological replicates that have been indicated with a small letter (“a”, “b” or “c”) after the number. In this study, biological replicates refer to samples removed from the same tube of sediment/soil or to a “twin” sterile swab used to swab the same portion of speleothem surface during the cave expeditions but that have been extracted and sequenced in the labs at different moments in time.
DNA, from a total of 21 sediment samples, was extracted in the metagenomics laboratory of Pusan National University (PNU) from 500 mg of sediment using FastDNA SPIN Kit for soil (MP Biomedicals, United States) following the manufacturer’s indication. The DNA has been quantified using a ThermoFisher Qubit-4 fluorometer and the purity defined by ThermoFisher Nanodrop spectrophotometer. 19 swab samples were sent to Zymo Research Corp., Irvine (CA, USA) laboratories and DNA extracted using ZymoBIOMICS-96 MagBead DNA Kit (Zymo Research, Irvine, CA).
2.3. Long-read sequencing
The DNA extracted in the PNU lab have been sequenced using the ONT MinION sequencer. Prior to the library preparation all samples have undergone an additional AMPure XP magnetic beads purification to increase the 260/230 values and remove contaminants, such as ethanol, that have been used during the DNA extraction step. For each sample, at least 1 μg of DNA has been used to prepare the sequencing library with the PCR-free SQK-LSK109 ligation kit (ONT) according to the manufacturer’s instructions. The libraries were loaded on FLO-MIN106 flow cells (R9.4.1 sequencing chemistry) inserted on the MinION device and basecalling was carried out using MinKNOW operating software (ONT) in fast basecalling mode.
In all cases, negative controls were included to assess the level of bioburden during the wet-lab procedures. We have tried to replicate each samples using both the technologies but due to low DNA yield during the extraction step, for some samples this has not been possible.
MinION raw reads’ barcodes were trimmed on both ends using the default filtering options of MinKNOWN (ONT) and the Qscore cut-off value was set to 7 or 9.
2.4. Short-read sequencing
The DNA extracted at Zymo Research Corp. laboratories were prepared for sequencing using Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA), that only requires reduced PCR cycles, with up to 100 ng DNA input and using internal dual-index 8 bp barcodes with Nextera adapters. The platform used for sequencing was NovaSeq 6000 (Illumina, San Diego, CA).
Illumina raw paired-end mode (150 nt) sequence reads were manually trimmed using Trimmomatic-0.33 (Bolger et al., 2014) to remove adapters: 6 bp window size for the trimming and a quality cutoff of 20. Reads with size lower than 70 bp were removed. Overrepresented G-poly-mers and duplicate reads were removed using bbduk and seqkit, respectively (Shen et al., 2016; Bushnell et al., 2017). Paired-end reads were merged using Pear (Zhang et al., 2014).
2.5. Bioinformatics analysis
Both Illumina and MinION cleaned and filtered reads were classified using 1.18.1 Kaiju (Menzel et al., 2016) in Greedy mode with 3 mismatches allowed against a subset of NCBI BLAST nr database (downloaded in April 2021) containing all proteins belonging to known Archaea, Bacteria, Viruses, Fungi, and some microbial Eukaryotes.
After the alignment to the database, the reads were filtered to remove reads with ambiguous taxa annotations and to exclude taxa with a mean less than 1 (counts) across all samples.
Read counts were processed for statistical analyses using functions in Phyloseq 4.2 R package (McMurdie and Holmes, 2013). Alpha diversity measures were carried out on clean and filtered data using Shannon index. Beta diversity was studied using non-metric multidimensional scaling (NMDS) and applying the Bray-Curtis distance using vegan R-packages.
3. Results
3.1. MinION samples taxonomic composition
The taxonomical classification led to a mean of 55% of successfully classified reads (Table 1). After filtering for taxa less than 2% to reduce the chances of false positive predictions, the total number of taxa decreased from 37,498 to 15,377. The taxonomy results were then normalized to 100%.
Among the taxa accounting for more than 2% of abundance, the most prominent ones are Proteobacteria and Actinobacteria (also known as Actinomycetes or Actinomycetota) followed by Acidobacteria, Planctomycetes, Chloroflexi, Candidatus Rokubacteria and Verrucomicrobia (Fig. 2). The microbial distributions of each cave are comparable. These communities are predominantly formed by Proteobacteria accounting for 36% and 33% and 30% of the total for Ondal, Sanjidang and Simbok Caves respectively. Actinobacteria presence along Ondal and Sanjidang Caves maintains stable on average at 13% and 9% respectively but in the samples taken near the entrance (ON01) and in deep sectors (ON8, ON11.a and ON11.b) it increases to on average 30% and 18% respectively. Similarly, Simbok Cave hosts relatively more Actinobacteria 21% of the total microbial composition. On a species level this phylum is dominated by a generic species of Actinobacteria and there is not a clear distinction between the three caves, although Ondal wet Cave shows a slight increase of the species Pseudonocardiales with respect to dry and seasonally wet caves (Fig 3a). Acidobacteria are a minor phylum: ranging from 12% in Ondal Cave, 13% in Simbok Cave and 15% in Sanjidang Cave.
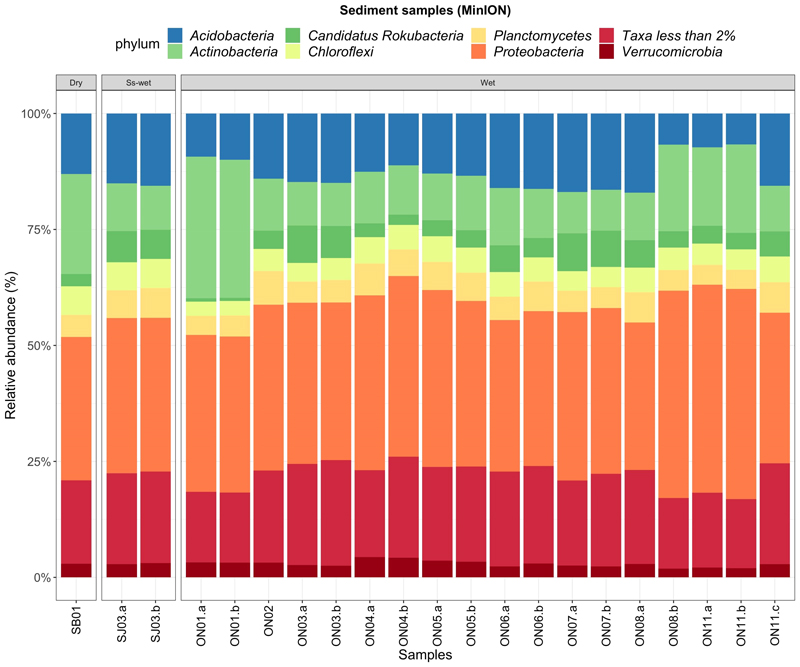
Bar-plot of the relative abundances of the most common phyla found in the different caves and sequenced using MinION Nanopore technology. The graph below the bar-plot shows the ratio between the major two phyla: Proteobacteria and Actinobacteria. The samples are grouped by type of environment: dry, seasonally wet (“Ss-wet”) and permanently wet (“Wet”) caves and the soil. SB = Simbok Cave, SJ = Sanjidang Cave, ON = Ondal Cave.
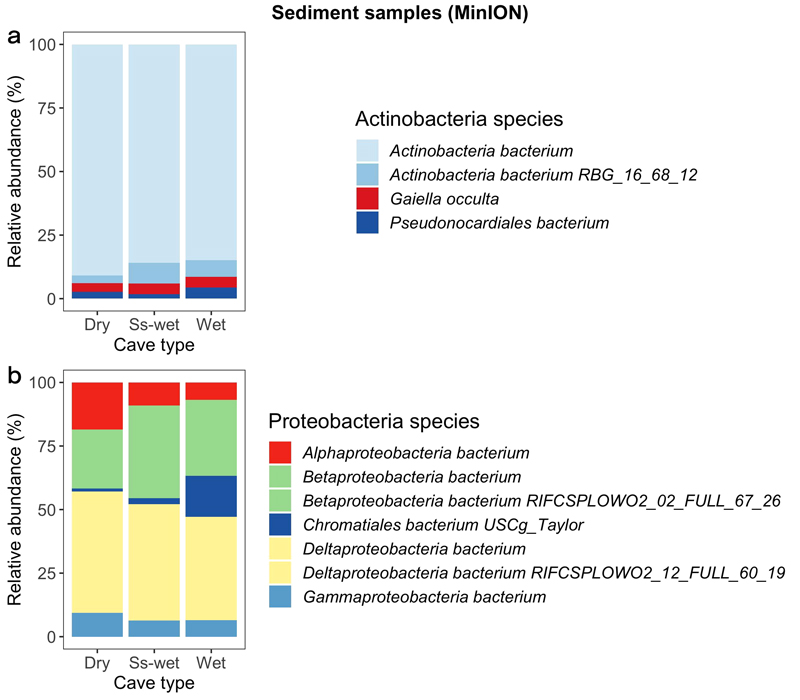
Compositional barplots normalized to 100% of main Proteobacteria species and Actinobacteria species from inside the caves grouped into dry, seasonally wet (Ss-wet) and wet groups and sequenced with nanopore MinION. Soil samples have been excluded from these plots. The two species of Actinobacteria bacterium, Betaproteobacteria bacterium and Deltaproteobacteria bacterium have been assigned with same colors (pale blue, green and yellow) because they represent generic species of that specific phylum. (Taxa filtering is based on a mean relative abundance greater than 15% and 30% for Actinobacteria species and Proteobacteria species respectively).
Proteobacteria is the dominant phylum found across all samples (Fig. 2). Among this phylum the most dominant species belong to the class of Deltaprotobacteria (also known as Mixococcota) and Betaprotobacteria (Fig. 3b). Betaprotobacteria show an increase in seasonally-wet and wet environments as well as the Gammaproteobacteria belonging to the Chromatiales species. On the contraty, Alphaproteobacteria are more abundant in dry environments compared to seasonally-wet and wet environments. Inside Ondal Cave sediment samples were collected from different locations (Fig. 4). Near the entrance of Ondal Cave, Betaprotobacteria and Alphaproteobacteria species are higher compared to other sites of the cave. Deep sections are mainly composed by the species Chromatiales and species of Deltaproteobacteria whereas species of Betaprotobacteria show a decrease compared to sectors near the entrance.
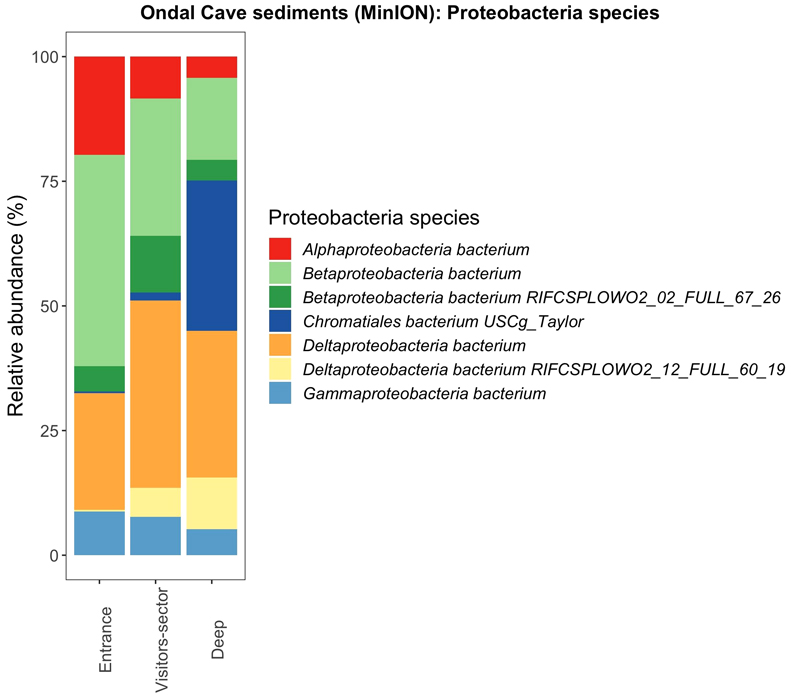
Compositional bar plots normalized to 100% of main Proteobacteria species for the samples from Ondal Cave sequenced with nanopore MinION. Samples are grouped depending on the site inside the cave where they have been collected: entrance, visitors sector and deep sector. The two species of Betaproteobacteria bacterium and Deltaproteobacteria bacterium have been assigned with same colors (green and yellow) because they represent generic species of that specific phylum. (Taxa filtering is based on a mean relative abundance greater than 30%).
3.2. Illumina samples taxonomic composition
The taxonomical classification led to a mean of 55% of successfully classified reads (Table 1). After filtering for taxa less than 2% to reduce the chances of false positive predictions, the total number of taxa decreased from 49,917 to 34,778. The taxonomy results were then normalized to 100%.
The swabs, like the sediment samples sequenced using the Nanopore MinION (section 3.1), mostly show the presence of Proteobacteria and Actinobacteria phyla (Fig. 5) showing an almost equal distribution of these two phyla. In Baram Cave samples, the dominant phylum is Actinobacteria accounting on average for the 65% of the total, whereas Protobacteria only accounts for the 11%. In Ondal Cave, Actinobacteria and Protobacteria abundances are comparable, being on average 35% and 34% of the total respectively. In Seodae and Sanjidang Caves, half of their microbial composition is formed by Proteobacteria, and only 13% on average is formed by Actinobacteria. The third most abundant phylum, Acidobacteria, has a constant abundance of less than 10% among all the swab samples from all the caves.
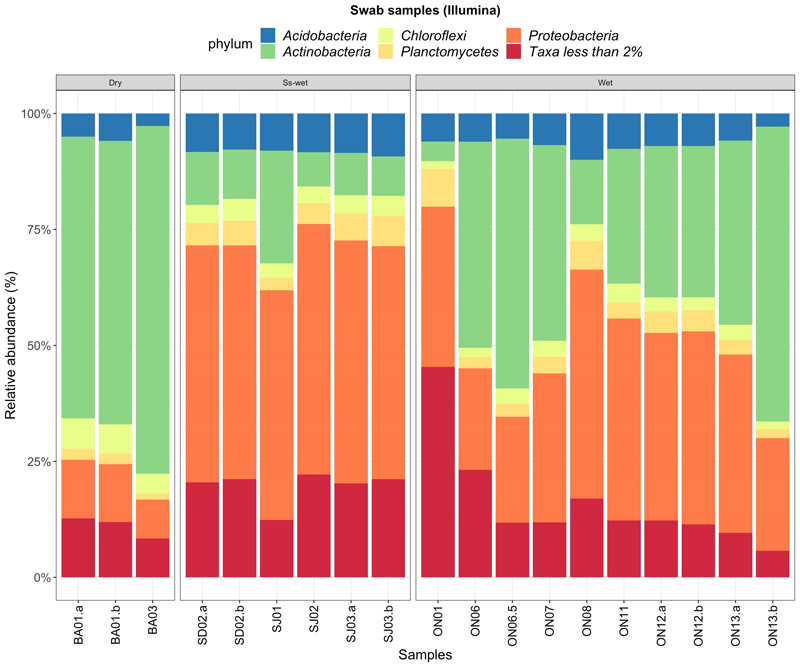
Bar-plot of the relative abundances of the most common phyla found in the different caves and sequenced using with Illumina NovaSeq. The graph below the bar-plot shows the ratio of the major two phyla: Proteobacteria and Actinobacteria, grouped by type of environment: dry, seasonally wet (“Ss-wet”) and permanently wet (“Wet”) caves and the soil. BA = Baram Cave, SD = Seodae Cave, SJ = Sanjidang Cave, ON = Ondal Cave.
A clear distinction in the composition of Proteobacteria and Actinobacteria species is noticeable between dry caves compared to wet and seasonally-wet caves (Fig. 6). Wet and seasonally-wet cave swab colonies mainly host the species Chromatiales (Phylum Gammaproteobacteria), whereas dry cave samples are mostly composed by species of Beta- and Deltaproteobacteria (Fig. 6b).
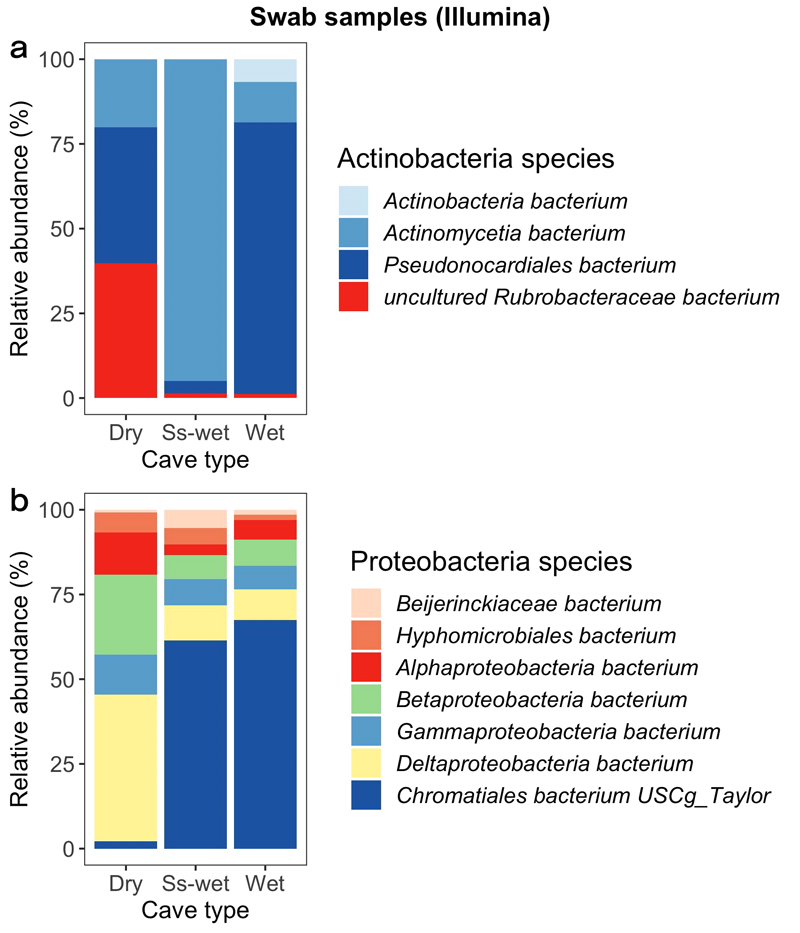
Compositional barplots normalized to 100% of main Proteobacteria species and Actinobacteria species (using cutoff at average abundance of 0.55%) from inside the caves grouped into dry, seasonally wet (Ss-wet) and wet groups and sequenced with Illumina NovaSeq. (Taxa filtering is based on a mean relative abundance greater than 30% for both Actinobacteria species and Proteobacteria species).
Regarding Actinobacteria, all the three categories of caves are different (Fig. 6a). Wet caves mostly host the species Pseudocardiales, for the 75% of the total abundance. Seasonally-wet cave specimen are almost entirely composed by the species Actinomycetia, while the dry caves present a more heterogenous microbial composition formed by the same amount (40%) of Pseudonocardiales and Rubrobacteraceae species and the rest by the species Actinomycetia.
Inside Ondal Cave sediment samples have been collected from different locations (Fig. 7). Near the entrance of the cave and in the sector where visitors are allowed in, the microbial composition is heterogeneous and mainly formed by species of Delta-, Alpha- and Proteobacteria. Deep sections are mainly dominated by the species Chromatiales.
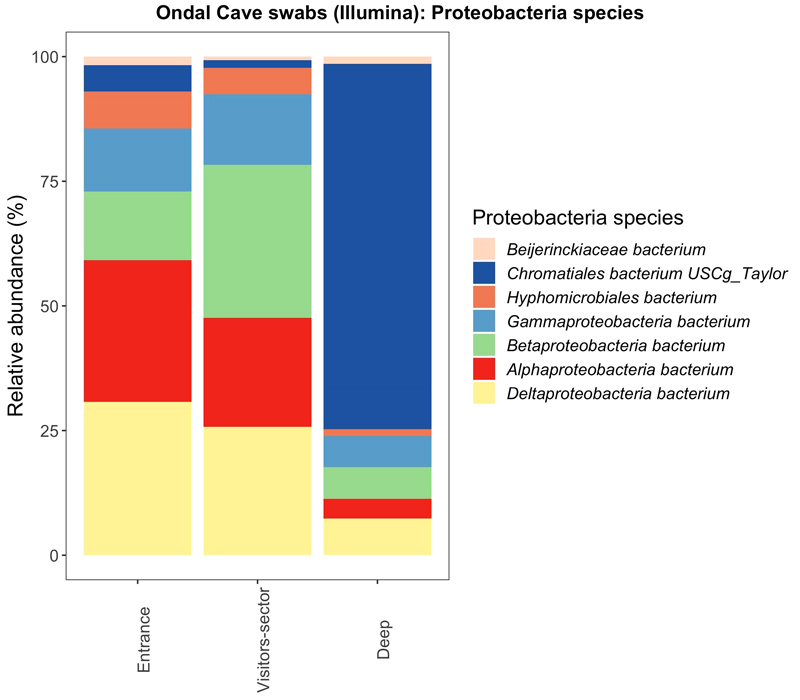
Compositional bar plots normalized to 100% of main Proteobacteria species for the samples from Ondal Cave sequenced with Illumina NovaSeq. Samples are grouped depending on the site inside the cave where they have been collected: entrance, visitors sector and deep sector. (Taxa filtering is based on a mean relative abundance greater than 30%).
3.3. Alpha diversity
The Illumina NovaSeq platform generated an average of 27,320,423 raw reads per sample compared to nanopore sequencing that produced an average of 437,510 raw reads per sample (Table 1 and 2). The ONT technology can analyze long fragments of DNA therefore, whereas the DNA sequenced with the NovaSeq had a fixed length of 150 bp, MinION reads have different lengths and their N50 values (weighted median) range from 1991 bp to 6100 bp (Table 1).
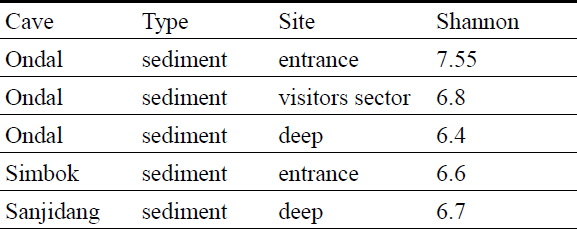
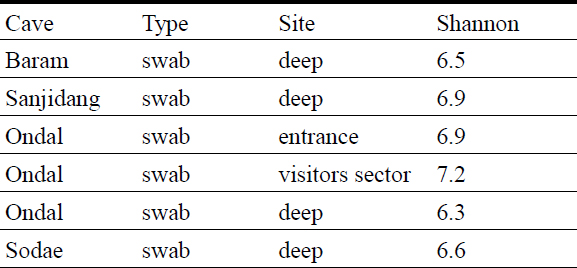
The diversity index was calculated on cleaned and filtered data. Considering the different sequencing depths obtained with the two different sequencing technologies, alpha diversity measures are discussed separately.
In general, the communities analyzed using Illumina Novaseq show similar diversity values of the communities analyzed with the MinION (Table 3 and 4). In Ondal Cave, deep-room communities have lower values compared to the entrance. The Shannon values from this study are higher compared to the values from another South Korean cave that ranged between 3 and 4.9 (Park et al., 2020), however they are similar to the values of cave microbiomes described by Ortiz et al. (2013) (USA) and Wiseschart et al. (2019) (Thailand), Addesso et al. (2021) (Italy).
3.4. Beta diversity
Ondal Cave’s bacterial communities grouped by sites: entrance, visitors sector and deep sector, were compared among each other to determine their similarity using non-metric multidimensional scaling (NMDS) (Supplementary Fig. 1). For the samples sequenced with MinION, the ordination analyses show three distinct groups of samples. Even though the samples collected near the entrance well separate from the other samples, 4 out of 7 samples collected in the deep sections of the cave fall within the group of the samples collected in the visitor sector. Illumina samples show three distinct groups in the NMDS space, and the samples cluster depending on the cave site where they were collected.
4. Discussion
This study aimed at testing whether cave-dwelling microbial communities are sensitive to environmental conditions inside South Korean caves, given their restricted availability of nutrients and the type of substrate. Therefore, since the main purpose of this study is to describe the biodiversity of the samples as a function of their surrounding environment, shotgun sequencing can be more appropriate compared to the 16S sequencing approach.
Shotgun sequencing, in contrast to the 16S ribosomal RNA sequencing method, can classify down to the species level while also aiming to estimate the relative abundances of the taxa within a sample (Tovo et al., 2020). Other advantages of the shotgun method compared to the 16S-amplicons approach include the mitigation of PCR biases, since it does not rely on the amplification of specific DNA target regions and the size of the database (Logares et al., 2014). In addition, the shotgun approach relies on larger and more comprehensive databases compared the amplicon homologue.
The choice of the Kaiju taxonomic classification algorithms has been reasoned on the proteins database used by the program, which are much more conserved with respect to nucleotide sequences (Menzel et al., 2016).
Cave bacterial consortia are influenced by the geology of the area (Zhu et al., 2019) and the geochemical characteristics of the surface but despite being geographically distant, the structure of the microbial communities from all these five South Korean caves is consistent with what is considered the core cave microbiome: Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, Chloroflexi, Planctomycetales, Bacteroidetes, Firmicutes, Acidobacteria, Actinobacteria, Nitrospirae, Gemmatimonadetes and Verrucomicrobia (Hershey and Barton, 2018) (Fig. 2 and 5). The source of these organisms is generally attributed to the soil microbiota transported inside the cave by vertical infiltration or surface streams, air currents or other animals (Kováč, 2018). Variations in the composition of microbial communities, both in the outside soil and in the dripping water, are also likely influenced by seasonal environmental parameters. Particularly Rasche et al. (2011) and Yun et al. (2016), showed that with high temperatures the abundance of Alphaproteobacteria, Acidobacteria and Verrucomicrobia decreases, whereas Betaproteobacteria increase. However, once in the cave, the selective pressure of this oligotrophic environment further sorts the bacterial species changing the structure of the original community of microbes (Ortiz et al., 2013; Lavoie et al., 2017) and even filtering contaminant species. The case of Lechuguilla Cave is emblematic as its native bacterial communities remained dominated by the endemic species despite being contaminated by humans’ urine commensal species introduced during some exploration trips (Johnston et al., 2012).
The most abundant bacteria in all the caves studied in this work are Proteobacteria and Actinobacteria, which are also the most common phyla found in many cave environments (Ortiz et al., 2013; Yun et al., 2016; de Mandal et al., 2017; Lavoie et al., 2017; Wiseschart et al., 2019; Zhu et al., 2019). Both phyla are ubiquitous in nature and Proteobacteria also constitute the largest and most diverse group of the Bacteria domain (Kersters et al., 2006).
Among the Proteobacteria phylum, bacteria belonging to the Gammaproteobacteria taxonomical class have been found in high percentages in the swab samples (Fig. 6 and 7). This agrees with Macalady et al. (2006)’s study, who found this type of Proteobacteria particularly dominant in biofilms in an Italian cave as well as with Jurado et al. (2020) work who mostly found Gammaproteobacteria in vermiculations from an Alpine cave. Similarly, in another cave in Slovenia, microbial communities from the walls were mostly dominated by members of Gammaproteobacteria (Pašić et al., 2009).
Proteobacteria have an important role in caves’ nitrogen cycle. Subphyla like Alpha- and Beta- Proteobacteria play critical role in the fixation of nitrogen, where N2 is fixated as ammonia (NH3) so that it can be utilized by other organisms, and they are also involved in the processes of nitrification and denitrification. Other subphyla like Gamma- and Delta- Proteobacteria are instead involved in other biogeochemical cycles, including sulfur and carbon cycles. They contribute to the carbon cycle by breaking down organic matter into simpler molecules that can be utilized by other microorganisms, or they take part in the process of sulphate reduction where sulphur gets recycled making it available for other biological processes, which is critical in anaerobic environment like caves. These processes are important in maintaining the balance of sulphur and nitrogen in cave ecosystems (Holmes et al., 1999; Ojeda et al., 2019; Knief et al., 2003; Macalady et al., 2006, 2008; Zhu et al., 2021, 2022). Some are also crucial in the formation of biofilms on cave surfaces (Engel et al., 2004) and in the processes of calcification of speleothems (Banks et al., 2010).
Actinobacteria have been found in association with pigment-forming communities on the walls of certain caves that are regularly visited by tourists suggesting that this phylum thrives in anthropized environments (Cuezva et al., 2012; Porca et al., 2012; Vautrin et al., 2021). Moreover, some of its species have also presented some pathogenic potential and health concerns for cave visitors (Jurado et al., 2010). Other species instead dominate pristine caves taking advantage of their ability of degrading complex compounds present in the cave, such as in nitrate reduction processes or to fix CO2 (Severino et al., 2019). Cave Actinobacteria, are also considered a potential source of novel bioactive compounds (Axenov-Gibanov et al., 2016; Adam et al., 2018).
The sediment species communities from all the caves in this study, show an increase of Acidobacteria species (Fig. 2) compared to the swabs where their presence is minimal (Fig. 5). This is a very common phylum of bacteria usually present in soils low in nutrients and often observed associated to Proteobacteria (Kielak et al., 2016).
At a species level, the most dominant taxa found on the walls of the caves from this study are Chromatiales (Gammaproteobacteria), Pseudonocardiales and Actinomycetia (Fig. 6 and 7). Similarly, the communities belonging to sediments from remote parts of the caves, showed an increase in Chromatiales Gammaproteobacteria (Fig. 4).
4.1. Distribution patterns of the major species in the cave niches
Chromatiales (Gammaproteobacteria), also known as purple sulfur bacteria, have been found to be one of the core microbes in many other cave environments around the world, such as in South Korea (Park et al., 2020), Spain, Czech Republic and Slovenia (Porca et al., 2012), Italy (D’Angeli et al., 2019), Brazil (Marques et al., 2019) and in an Azorean lava cave (Riquelme et al., 2015). Despite it does not surprise to find this species on wet rock surfaces (Imhoff, 2006), these are phototrophic organisms hence light is a selective factor for their development (Rosenberg et al., 2006). It is, therefore, unexpected to find Chromatiales at high percentages in deep sections of Ondal, Seodae and Sanjidang Caves that are not illuminated by natural nor artificial lights (Fig. 4 and 7). A similar result occurred to Marques et al. (2019), who found Chromatiales dominating the bacterial communities in aphotic zones of a Brazilian cave and interpreted its presence for being associated to the nitrogen cycle and ammonia oxidation processes. Nitrogen-based metabolism is indeed commonly found in bacteria adapted to environments scarce in nutrients (Ortiz et al., 2014; Reboleira et al., 2022). Zhu et al. (2019) also presumed that nitrogen is an important source of nutrients for microbes living in caves and that can decrease the overall species richness in favor of only certain dominant taxa, like Chromatiales indeed. This is consistent with soil cover above karst systems being generally saturated in N (Wen et al., 2016; Chen et al., 2019) that can leach inside the cave through seepage. Therefore, the presence of this species that have been found in seasonally-wet and wet caves could be mainly related to relatively high humid levels in the environment and nitrogen availability.
The other dominant species is Pseudonocardiales bacterium (Actinobacteria), found at relatively high percentages in the swab communities from Ondal and Baram Caves (Fig. 6), which are wet and dry caves respectively despite being very low in all the sediment samples (Fig. 3). This species of bacteria is commonly found in caves, and it has been proposed as being an indicator of specialized limestone bacterial communities (Zhu et al., 2019) and it was also found to be the major constituent in microbial biofilms covering Paleolithic paintings in some Spanish caves (Stomeo et al., 2008).
The presence of members belonging to Pseudonocardiaceae has been described as part of the core microbiome of an unmarked cave environment (Porca et al., 2012; Riquelme et al., 2015; Lavoie et al., 2017; Buresova-Faitova et al., 2022). In that regard, we can understand the increased counts of Pseudonocardiales bacterium in the undisturbed Baram Cave or in the non-touristic sectors of Ondal Cave (Fig. 6). Despite Seodae and Sanjidang being both pristine caves, the presence of this species of Actinobacteria is very low. This could be interpreted because these caves mostly host species of Proteobacteria, at the expenses of Actinobacteria, that are linked to consistent availability of Dissolved Organic Carbon (DOC) and high humidity. This order of bacteria, when living in the soil, degrades aromatic compounds and converts organic litter into humic acids. By transporting aromatic compounds, drip waters contribute to feed and sustain the community of this type of bacteria inside caves (Marques et al., 2019; Turrini et al., 2020). The same can be said for Ondal Cave that in terms of water availably is more similar to Seodae and Sajindang Caves compared to Baram cave.
Actinobacteria have also been interpreted as indicators of bare dry rock surfaces or inactive speleothems (Vardeh et al., 2018). Considering that Baram Cave is also high in other Actinobacteria species, such as Rubrobacteraceae bacterium (Fig. 6), this microorganisms could be considered as a proxy of a dry cave environment. The cave was indeed ventilated and dry at the time of the visit, with no speleothem formations and the bare bedrock is exposed for all its length. Crucially, Rubrobacterales, is an order of bacteria highly desiccation-resistant that generally grows after an active microbial mat passes to be inactive (Vardeh et al., 2018). This also agrees with Baram Cave relatively lower abundance of Proteobacteria that are associated to highly available organic matter and relatively wet and humid environments.
Cave walls, compared to lose and porous sediments, are better substrates to form biofilms and bacterial colonies where dissolved nutrients from the topsoil can be provided by dripping water as demonstrated by the presence in the swabs of cave wall dweller species such as Hyphomicrobiales (Alpha-Proteobacteria) (Jurado et al. 2022) and Actinomycetia (Groth and Saiz-Jimenez, 1999; Pašić et al., 2009; Farda et al., 2022), which are absent in the sediments.
In general, it is possible to notice that the distribution of the phyla in the two types of substrates is different (Fig. 2 and 5). In the sediments, the phyla abundances are more constantly distributed compared to the swabs where they fluctuate more likely reflecting more specialized colonies due to limited nutrients input from the outside environment (Zhu et al., 2019).
In addition, examining the distribution of microbes within cave surfaces like stalagmite or flowstone layers provides an additional method to interpret paleoclimate data obtained from speleothem archives. Embracing a multidisciplinary scientific approach to climate science, metagenomics could complement the analysis of stable isotope records (18O and 13C), trace element distribution and the petrographic study of crystalline fabrics (Fairchild and Baker, 2012).
5. Conclusions
The microbial composition identified for the South Korean caves investigated in this study agrees with the core microbiome for karstic environments (Hershey and Barton, 2018). The more common bacteria found in these colonies belong to the phyla Actinobacteria and Proteobacteria. In general, Proteobacteria dominate communities highly exposed to nutrients and animals. Natural sources of organic matter in caves are DOC and colloidal organics from the soil layers above, then transported by subterranean waters, either dripping or floodings. The second source of nutrients comes from plants and animals (Ikner et al., 2007). Proteobacteria that comprise Gamma-, Beta-, Alpha- and Deltaproteobacteria classes, are largely widespread thanks to their capacity of degrading a broad spectrum of organic materials and thus to survive in hostile and nutrients-deprived environments, like caves (Addesso et al., 2021). In either way, Protobacteria not only are associated to high source of organic matter but also to relatively wet and humid environments where nutrients can be easily transported around (Pašić et al., 2009).
In our study we found high abundances of Proteobacteria for all the well decorated caves with active dripping like in the case of Ondal Cave or seasonally wet, during the summer monsoons, caves like Sanjidang and Seodae Caves (Fig. 2 and 5). The only caves with relatively low percentages of Proteobacteria are Baram and Simbok Caves that were dry and dusty during the exploration and lack speleothem deposits (Fig. 2 and 5). The major pattern of Proteobacteria distribution observed in these caves is a relative increase of Gammaproteobacteria in the deep sectors contrary to Alpha- and Beta-Proteobacteria that show a decrease and can be potentially interpret as due to a major nitrogen availability in these parts of the caves, that would need to be confirmed with a metabolic pathways study. A major presence of Actinobacteria relative to Proteobacteria is here interpreted as a proxy of a relatively dry environment, like in the case of Baram (Fig. 5) and Simbok (Fig. 2) Caves. Whether the relative abundance of Rubrobacteraceae and Chromatiales species of bacteria can be regarded as proxy for dry and wet cave conditions respectively, needs to be further explored in the future. In summary, specific microhabitats play a crucial role in controlling the distribution of microbial communities even in the face of an overall resemblance in microbial communities amid geographically distant cave systems.
Data availability statement
The datasets generated and/or analysed in this study are available in the Sequence Read Archive (SRA), hosted by the National Center for Biotechnology Information (NCBI), available with accession number PRJNA946675.
Acknowledgments
This research was supported by the Institute for Basic Science (IBS) IBS-R028-D1. The kaiju taxonomic classification of sequencing reads was partly performed on the IBS/ICCP supercomputer “Aleph,” 1.43 peta flops high-performance Cray XC50-LC Skylake computing system with 18,720 processor cores, 9.59 PB storage, and 43 PB tape archive space. We also acknowledge the support of KREONET. The cave DNA samples were obtained under the license permission number 2023-0023 by the Cultural Heritage Administration of the Republic of Korea. We would like to thank Prof. Jihwan Hwang from the Anti-Microbial Control Laboratory at Pusan National University for the Nanodrop analyses.
References
-
Adam, D., Maciejewska, M., Naome, A., Martinet, L., Coppieters, W., Karim, L., Baurain, D. and Rigali, S., 2018, Isolation, characterization, and antibacterial activity of hard-to-culture Actino-bacteria from cave Moonmilk deposits. Antibiotics, 7, 28.
[https://doi.org/10.3390/antibiotics7020028]

-
Addesso, R., Gonzalez-Pimentel, J.L., D’angeli, I.M., De Waele, J., Saiz-Jimenez, C., Jurado, V., Miller, A.Z., Cubero, B., Vigliotta, G. and Baldantoni, D., 2021, Microbial Community Characterizing Vermiculations from Karst Caves and Its Role in Their Formation. Environmental Microbiology, 81, 884-896.
[https://doi.org/10.1007/s00248-020-01623-5]

-
Axenov-Gibanov, D.V., Voytsekhovskaya, I.V., Tokovenko, B.T., Protasov, E.S., Gamaiunov, S.V., Rebets, Y.V., Luzhetskyy, A.N. and Timofeyev, M.A., 2016, Actinobacteria isolated from an underground Lake and Moonmilk speleothem from the biggest conglomeratic karstic cave in Siberia as sources of novel biologically active compounds. PLoS One, 11, e0149216.
[https://doi.org/10.1371/journal.pone.0149216]

-
Banks, E.D., Taylor, N.M., Gulley, J., Lubbers, B.R., Giarrizzo, J.G., Bullen, H.A., Hoehler, T.M. and Barton, H.A., 2010, Bacterial calcium carbonate precipitation in cave environments: a function of calcium homeostasis. Geomicrobiology Journal, 27, 444-454.
[https://doi.org/10.1080/01490450903485136]

- Barton, H.A., 2006, Introduction to cave microbiology: A review for the non-specialist. Journal of Cave and Karst Studies, 68, 43-54.
-
Bolger, A.M., Lohse, M. and Usadel, B., 2014, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114-2120.
[https://doi.org/10.1093/bioinformatics/btu170]

-
Buresova-Faitova, A., Kopecky, J., Sagova-Mareckova, M., Alonso, L., Vautrin, F., Moënne-Loccoz, Y. and Rodriguez-Nava, V., 2022, Comparison of Actinobacteria communities from human-impacted and pristine karst caves. Microbiology Open, 11, e1276.
[https://doi.org/10.1002/mbo3.1276]

-
Bushnell, B., Rood, J. and Singer, E., 2017, BBMerge - Accurate paired shotgun read merging via overlap. PLoS One, 12, e0185056.
[https://doi.org/10.1371/journal.pone.0185056]

-
Cacchio, P., Contento, R., Ercole, C., Cappuccio, G., Martinez, M.P. and Lepidi, A., 2004, Involvement of microorganisms in the formation of carbonate speleothems in the Cervo cave (L’Aquila-Italy). Geomicrobiology Journal, 21, 497-509.
[https://doi.org/10.1080/01490450490888109]

-
Cacchio, P., Ercole, C., Cappuccio, G. and Lepidi, A., 2003, Calcium carbonate precipitation by bacterial strains isolated from a limestone cave and from a loamy soil. Geomicrobiology Journal, 20, 85-98.
[https://doi.org/10.1080/01490450303883]

-
Carmichael, M.J., Carmichael, S.K., Santelli, C.M., Strom, A. and Brauer, S.L., 2013, Mn(II)-oxidizing Bacteria are Abundant and Environmentally Relevant Members of Ferromanganese Deposits in Caves of the Upper Tennessee River Basin. Geomicrobiology Journal 30, 779-800.
[https://doi.org/10.1080/01490451.2013.769651]

-
Chang, G.D., Kim, J.W., Lee, T.K., Beak, S.A., Choi, Y.G., Park, K.H. and Joo, S., 2021, Biota of limestone caves in Pyeongchang-gun, Korea: Focusing on three undisclosed caves. Journal of Asia-Pacific Biodiversity, 14, 302-312.
[https://doi.org/10.1016/j.japb.2021.05.003]

-
Chen, H., Li, D., Mao, Q., Xiao, K. and Wang, K., 2019, Resource limitation of soil microbes in karst ecosystems. Science of the Total Environment, 650, 241-248.
[https://doi.org/10.1016/j.scitotenv.2018.09.036]

-
Chiang, A.D. and Dekker, J.P., 2020, From the Pipeline to the Bedside: Advances and Challenges in Clinical Metagenomics. The Journal of Infectious Diseases, 221, S331-S340.
[https://doi.org/10.1093/infdis/jiz151]

- Choi, D.W., Woo, K.S., Kim, R. and Lee, K.C., 2003, The origin of the Okgye Cave and the genesis of spleothems, Kangneung, Korea. Journal of the Geological Society of Korea, 39, 41-64 (in Korean with English abstract).
-
Ciuffreda, L., Rodríguez-Pérez, H. and Flores, C., 2021, Nanopore sequencing and its application to the study of microbial communities. Computational and Structural Biotechnology Journal, 19, 1497-1511.
[https://doi.org/10.1016/j.csbj.2021.02.020]

-
Cuezva, S., Fernandez-Cortes, A., Porca, E., Pašić, L., Jurado, V., Hernandez-Marine, M., Serrano-Ortiz, P., Hermosin, B., Cañaveras, J.C., Sanchez-Moral, S. and Saiz-Jimenez, C., 2012, The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiology Ecology, 81, 281-290.
[https://doi.org/10.1111/j.1574-6941.2012.01391.x]

-
D’Angeli, I.M., Ghezzi, D., Leuko, S., Firrincieli, A., Parise, M., Fiorucci, A., Vigna, B., Addesso, R., Baldantoni, D., Carbone, C., Miller, A.Z., Jurado, V., Saiz-Jimenez, C., de Waele, J. and Cappelletti, M., 2019, Geomicrobiology of a seawater-influenced active sulfuric acid cave. PLoS One, 14, e0220706.
[https://doi.org/10.1371/journal.pone.0220706]

-
De Mandal, S., Chatterjee, R. and Kumar, N.S., 2017, Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiology, 17, 90.
[https://doi.org/10.1186/s12866-017-1002-x]

-
Dimkić, I., Fira, D., Janakiev, T., Kabić, J., Stupar, M., Nenadić, M., Unković, N. and Ljaljević Grbić, M., 2021, The microbiome of bat guano: for what is this knowledge important?. Applied Microbiology and Biotechnology, 105, 1407-1419.
[https://doi.org/10.1007/s00253-021-11143-y]

-
Dong, Y., Gao, J., Wu, Q., Ai, Y., Huang, Y., Wei, W., Sun, S. and Weng, Q., 2020, Co-occurrence pattern and function prediction of bacterial community in Karst cave. BMC Microbiology, 20, 137.
[https://doi.org/10.1186/s12866-020-01806-7]

-
Engel, A.S., Porter, M.L., Stern, L.A., Quinlan, S. and Bennett, P.C., 2004, Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria”. FEMS Microbiology Ecology, 51, 31-53.
[https://doi.org/10.1016/j.femsec.2004.07.004]

-
Fairchild, I. and Baker, A., 2012, Speleothem Science: From Process to Past Environments. John Wiley & Sons.
[https://doi.org/10.1002/9781444361094]

-
Farda, B., Djebaili, R., Vaccarelli, I., Del Gallo, M. and Pellegrini, M., 2022, Actinomycetes from caves: an overview of their diversity, biotechnological properties, and insights for their use in soil environments. Microorganisms, 10, 453.
[https://doi.org/10.3390/microorganisms10020453]

-
Franchi, F. and Frisia, S., 2020, Crystallization pathways in the Great Artesian Basin (Australia) spring mound carbonates: Implications for life signatures on Earth and beyond. Sedimentology, 67, 2561-2595.
[https://doi.org/10.1111/sed.12711]

-
Ghosh, S., Kuisiene, N. and Cheetham, N., 2017, The cave microbiome as a source for drug discovery: Reality or pipe dream?. Biochemical Pharmacology, 134, 18-34.
[https://doi.org/10.1016/j.bcp.2016.11.018]

-
Graening, G.O. and Brown, A.V., 2003, Ecosystem dynamics and pollution effects in an Ozark cave stream. Journal of the American Water Resources Association, 39, 1497-1507.
[https://doi.org/10.1111/j.1752-1688.2003.tb04434.x]

-
Hershey, O.S. and Barton, H.A., 2018, The Microbial Diversity of Caves. In: Moldovan, O.T., Kováč, Ľ., Halse, S. (Eds.), Cave Ecology. Springer International Publishing, Cham, 69-90.
[https://doi.org/10.1007/978-3-319-98852-8_5]

-
Holmes, A.J., Roslev, P., McDonald, I.R., Iversen, N., Henriksen, K. and Murrell, J.C., 1999, Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Applied Environmental Microbiology, 65, 3312-3318.
[https://doi.org/10.1128/AEM.65.8.3312-3318.1999]

-
Ikner, L.A., Toomey, R.S., Nolan, G., Neilson, J.W., Pryor, B.M. and Maier, R.M., 2007, Culturable Microbial Diversity and the Impact of Tourism in Kartchner Caverns, Arizona. Microbiology Ecology, 53, 30-42.
[https://doi.org/10.1007/s00248-006-9135-8]

-
Imhoff, J.F., 2006, The Chromatiaceae. In: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E. (Eds.), The Prokaryotes: A Handbook on the Biology of Bacteria Volume 6, Proteobacteria: Gamma Subclass. Springer New York, New York, NY, 846-873.
[https://doi.org/10.1007/0-387-30746-X_31]

-
Jaroszewicz, W., Bielańska, P., Lubomska, D., Kosznik-Kwaśnicka, K., Golec, P., Grabowski, Ł., Wieczerzak, E., Dróżdż, W., Gaffke, L., Pierzynowska, K., Wegrzyn, G. and Wegrzyn, A., 2021, Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland). Antibiotics, 10, 1212.
[https://doi.org/10.3390/antibiotics10101212]

-
Jo, K., Woo, K.S., Hong, G.H., Kim, S.H. and Suk, B.C., 2010, Rainfall and hydrological controls on speleothem geochemistry during climatic events (droughts and typhoons): An example from Seopdong Cave, Republic of Korea. Earth and Planetary Science Letters, 295, 441-450.
[https://doi.org/10.1016/j.epsl.2010.04.024]

-
Jo, K., Woo, K.S., Yi, S., Yang, D.Y., Lim, H.S., Wang, Y., Cheng, H. and Edwards, R.L., 2014, Mid-latitude interhemispheric hydrologic seesaw over the past 550,000 years. Nature, 508, 378-382.
[https://doi.org/10.1038/nature13076]

-
Jo, K., Yi, S., Lee, J.-Y., Woo, K.S., Cheng, H., Edwards, L.R. and Kim, S.-T., 2017, 1000-Year Quasi-Periodicity of Weak Monsoon Events in Temperate Northeast Asia since the Mid-Holocene. Scientific Reports, 7, 15196.
[https://doi.org/10.1038/s41598-017-15566-4]

-
Johnston, M.D., Muench, B.A., Banks, E.D. and Barton, H.A., 2012, Human urine in lechuguilla cave: the microbiological impact and potential for bioremediation. Journal of Cave and Karst Studies, 74, 278-291.
[https://doi.org/10.4311/2011MB0227]

-
Jones, D.S., Schaperdoth, I. and Macalady, J.L., 2016, Biogeography of sulfur-oxidizing Acidithiobacillus populations in extremely acidic cave biofilms. ISME Journal, 10, 2879-2891.
[https://doi.org/10.1038/ismej.2016.74]

-
Jurado, V., Del Rosal, Y., Jimenez de Cisneros, C., Liñan, C., Martin-Pozas, T., Gonzalez-Pimentel, J.L., Hermosin, B. and Saiz-Jimenez, C., 2022, Microbial communities in carbonate precipitates from drip waters in Nerja Cave, Spain. PeerJ, 10, e13399.
[https://doi.org/10.7717/peerj.13399]

-
Jurado, V., Gonzalez-Pimentel, J.L., Miller, A.Z., Hermosin, B., D’Angeli, I.M., Tognini, P., de Waele, J. and Saiz-Jimenez, C., 2020, Microbial Communities in Vermiculation Deposits from an Alpine Cave. Frontiers in Earth Sciences, 8, 1-17.
[https://doi.org/10.3389/feart.2020.586248]

-
Jurado, V., Laiz, L., Rodriguez-Nava, V., Boiron, P., Hermosin, B., Sanchez-Moral, S. and Saiz-Jimenez, C., 2010, Pathogenic and opportunistic microorganisms in caves. International Journal of Speleology, 39, 15-24.
[https://doi.org/10.5038/1827-806X.39.1.2]

-
Kato, N.N., Arini, G.S., Silva, R.R., Bichuette, M.E., Bitencourtc, J.A.P. and Lopes, N.P., 2024, The World of Cave Microbiomes: Biodiversity, Ecological Interactions, Chemistry,and the Multi-Omics Integration. Journal of the Brazilian Chemical Society, 35, e-20230148.
[https://doi.org/10.21577/0103-5053.20230148]

-
Kersters, K., de Vos, P., Gillis, M., Swings, J., Vandamme, P. and Stackebrandt, E., 2006, Introduction to the Proteobacteria. In: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E. (Eds.), The Prokaryotes: Volume 5, Proteobacteria: Alpha and Beta Subclasses. Springer New York, New York, NY, 3-37.
[https://doi.org/10.1007/0-387-30745-1_1]

-
Kielak, A.M., Barreto, C.C., Kowalchuk, G.A., van Veen, J.A. and Kuramae, E.E., 2016, The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Frontiers in microbiology, 7, 744.
[https://doi.org/10.3389/fmicb.2016.00744]

-
Kim, D.S., Park, S.J., Han, S.H., Choi, D.W., Choi, Y.G., Kim, H.J., Kim, D.G. and Yi, H., 2018, Ecological characteristics of a cave-dwelling moth species, Triphosa dubitata (Lepidoptera: Geometridae), in Baram and Ssang caves in Gangwon Province, Korea. Entomological Research, 48, 92-98.
[https://doi.org/10.1111/1748-5967.12254]

-
Knief, C., Lipski, A. and Dunfield, P.F., 2003, Diversity and activity of Methanotrophic bacteria in different upland soils. Applied Environmental Microbiology, 69, 6703-6714.
[https://doi.org/10.1128/AEM.69.11.6703-6714.2003]

-
Kosznik-Kwaśnicka, K., Golec, P., Jaroszewicz, W., Lubomska, D. and Piechowicz, L., 2022, Into the Unknown: Microbial Communities in Caves, Their Role, and Potential Use. Microorganisms, 10, 222.
[https://doi.org/10.3390/microorganisms10020222]

-
Kováč, L., 2018, Caves as Oligotrophic Ecosystems. In: Moldovan, O.T., Kováč, Ľ., Halse, S. (Eds.), Cave Ecology. Springer International Publishing, Cham, 297-307.
[https://doi.org/10.1007/978-3-319-98852-8_13]

-
Lavoie, K.H., Winter, A.S., Read, K.J.H., Hughes, E.M., Spilde, M.N. and Northup, D.E., 2017, Comparison of bacterial communities from lava cave microbial mats to overlying surface soils from Lava Beds National Monument, USA. PLoS One, 12, e0169339.
[https://doi.org/10.1371/journal.pone.0169339]

-
Lee, C.W., Tomikawa, K.O., Nakano, T. and Min, G.S., 2020, A new species of the genus Pseudocrangonyx (Crustacea: Amphipoda: Pseudocrangonyctidae) from Simbok Cave, Korea. Zootaxa, 4731, 321-334.
[https://doi.org/10.11646/zootaxa.4731.3.2]

-
Logares, R., Sunagawa, S., Salazar, G., Cornejo-Castillo, F.M., Ferrera, I., Sarmento, H., Hingamp, P., Ogata, H., de Vargas, C., Lima-Mendez, G., Raes, J., Poulain, J., Jaillon, O., Wincker, P., Kandels-Lewis, S., Karsenti, E., Bork, P. and Acinas, S.G., 2014, Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environmental Microbiology, 16, 2659-2671.
[https://doi.org/10.1111/1462-2920.12250]

-
Macalady, J.L., Dattagupta, S., Schaperdoth, I., Jones, D.S., Druschel, G.K. and Eastman, D., 2008, Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. The ISME Journal, 2, 590-601.
[https://doi.org/10.1038/ismej.2008.25]

-
Macalady, J.L., Lyon, E.H., Koffman, B., Albertson, L.K., Meyer, K., Galdenzi, S. and Mariani, S., 2006, Dominant Microbial Populations in Limestone-Corroding Stream Biofilms, Frasassi Cave System, Italy. Applied and Environmental Microbiology, 72, 5596-5609.
[https://doi.org/10.1128/AEM.00715-06]

-
Marques, E.L.S., Dias, J.C.T., Gross, E., Silva, A.B. de C. e, de Moura, S.R. and Rezende, R.P., 2019, Purple Sulfur Bacteria Dominate Microbial Community in Brazilian Limestone Cave. Microorganisms, 7, 29.
[https://doi.org/10.3390/microorganisms7020029]

-
Martin-Pozas, T., Fernandez-Cortes, A., Cuezva, S., Cañaveras, J.C., Benavente, D., Duarte, E., Saiz-Jimenez, C. and Sanchez-Moral, S., 2023, New insights into the structure, microbial diversity and ecology of yellow biofilms in a Paleolithic rock art cave (Pindal Cave, Asturias, Spain). Science of The Total Environment, 897, 165218.
[https://doi.org/10.1016/j.scitotenv.2023.165218]

-
McMurdie, P.J. and Holmes, S., 2013, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One, 8, e61217.
[https://doi.org/10.1371/journal.pone.0061217]

-
Menzel, P., Ng, K.L. and Krogh, A., 2016, Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communication, 7, 11257.
[https://doi.org/10.1038/ncomms11257]

-
Ortiz, M., Legatzki, A., Neilson, J.W., Fryslie, B., Nelson, W.M., Wing, R.A., Soderlund, C.A., Pryor, B.M. and Maier, R.M., 2014, Making a living while starving in the dark: Metagenomic insights into the energy dynamics of a carbonate cave. ISME Journal, 8, 478-491.
[https://doi.org/10.1038/ismej.2013.159]

-
Ortiz, M., Neilson, J.W., Nelson, W.M., Legatzki, A., Byrne, A., Yu, Y., Wing, R.A., Soderlund, C.A., Pryor, B.M., Pierson, L.S. and Maier, R.M., 2013, Profiling Bacterial Diversity and Taxonomic Composition on Speleothem Surfaces in Kartchner Caverns, AZ. Microbiology Ecology, 65, 371-383.
[https://doi.org/10.1007/s00248-012-0143-6]

-
Park, S., Cho, Y.J., Jung, D.Y., Jo, K.N., Lee, E.J. and Lee, J.S., 2020, Microbial Diversity in Moonmilk of Baeg-nyong Cave, Korean CZO. Frontiers in Microbiology, 11, 613.
[https://doi.org/10.3389/fmicb.2020.00613]

-
Parker, C.W., Senko, J.M., Auler, A.S., Sasowsky, I.D., Schulz, F., Woyke, T. and Barton, H.A., 2022, Enhanced terrestrial Fe(II) mobilization identified through a novel mechanism of microbially driven cave formation in Fe(III)-rich rocks. Scientific Reports, 12, 17062.
[https://doi.org/10.1038/s41598-022-21365-3]

-
Pašić, L., Kovče, B., Sket, B. and Herzog-Velikonja, B., 2009, Diversity of microbial communities colonizing the walls of a Karstic cave in Slovenia. FEMS Microbiology Ecology, 71, 50-60.
[https://doi.org/10.1111/j.1574-6941.2009.00789.x]

-
Porca, E., Jurado, V., Žgur-Bertok, D., Saiz-Jimenez, C. and Pašić, L., 2012, Comparative analysis of yellow microbial communities growing on the walls of geographically distinct caves indicates a common core of microorganisms involved in their formation. FEMS Microbiology Ecology, 81, 255-266.
[https://doi.org/10.1111/j.1574-6941.2012.01383.x]

-
Rasche, F., Knapp, D., Kaiser, C., Koranda, M., Kitzler, B., Zechmeister-Boltenstern, S., Richter, A. and Sessitsch, A., 2011, Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME Journal, 5, 389-402.
[https://doi.org/10.1038/ismej.2010.138]

-
Reboleira, A.S., Bodawatta, K.H., Ravn, N.M.R., Lauritzen, S.-E., Skoglund, R.Ø., Poulsen, M., Michelsen, A. and Jønsson, K.A., 2022, Nutrient-limited subarctic caves harbour more diverse and complex bacterial communities than their surface soil. Environmental Microbiome, 17, 41.
[https://doi.org/10.1186/s40793-022-00435-z]

-
Riquelme, C., Marshall Hathaway, J.J., Enes Dapkevicius, M.D.L., Miller, A.Z., Kooser, A., Northup, D.E., Jurado, V., Fernandez, O., Saiz-Jimenez, C. and Cheeptham, N., 2015, Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Frontiers in Microbiology, 6, 1342.
[https://doi.org/10.3389/fmicb.2015.01342]

-
Riquelme, C., Rigal, F., Hathaway, J.J.M., Northup, D.E., Spilde, M.N., Borges, P.A.V., Gabriel, R., Amorim, I.R. and Dapkevicius, M. de L.N.E., 2015, Cave microbial community composition in oceanic islands: disentangling the effect of different colored mats in diversity patterns of Azorean lava caves. FEMS Microbiology Ecology, 91, fiv141.
[https://doi.org/10.1093/femsec/fiv141]

-
Roldán, M. and Hernández-Mariné, M., 2009, Exploring the secrets of the three-dimensional architecture of phototrophic biofilms in caves. International Journal of Speleology, 38, 41-53.
[https://doi.org/10.5038/1827-806X.38.1.5]

- Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E. and Thompson, F., 2006, The Prokaryotes, 3rd ed. Springer New York, NY.
-
Severino, R., Froufe, H.J.C., Barroso, C., Albuquerque, L., Loboda-Cunha, A., da Costa, M.S. and Egas, C., 2019, High-quality draft genome sequence of Gaiella occulta isolated from a 150 meter deep mineral water borehole and comparisonwith the genome sequences of other deep-branching lineages of the phylum Actinobacteria. Microbiology Open, e840.
[https://doi.org/10.1002/mbo3.840]

-
Shen, W., Le, S., Li, Y. and Hu, F., 2016, SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One, 11, e0163962.
[https://doi.org/10.1371/journal.pone.0163962]

-
Stahlschmidt, M.C., Collin, T.C., Fernandes, D.M., Bar-Oz, G., Belfer-Cohen, A., Gao, Z., Jakeli, N., Matskevich, Z., Meshveliani, T., Pritchard, J.K., McDermott, F. and Pinhasi, R., 2019, Ancient Mammalian and Plant DNA from Late Quaternary Stalagmite Layers at Solkota Cave, Georgia. Scientific Reports, 9, 6628.
[https://doi.org/10.1038/s41598-019-43147-0]

-
Stomeo, F., Portillo, M.C., Gonzalez, J.M., Laiz, L. and Saiz-Jimenez, C., 2008, Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int Biodeterior Biodegradation, 62, 483-486.
[https://doi.org/10.1016/j.ibiod.2007.12.011]

-
Tomova, I., Lazarkevich, I., Tomova, A., Kambourova, M. and Vasileva-Tonkova, E., 2013, Diversity and biosynthetic potential of culturable aerobic heterotrophic bacteria isolated from Magura Cave, Bulgaria. International Journal of Speleology, 42, 65-76.
[https://doi.org/10.5038/1827-806X.42.1.8]

-
Tovo, A., Menzel, P., Krogh, A., Lagomarsino, M.C. and Suweis, S., 2020, Taxonomic classification method for metagenomics based on core protein families with Core-Kaiju. Nucleic Acids Research, 48, 93.
[https://doi.org/10.1093/nar/gkaa568]

-
Turrini, P., Tescari, M., Visaggio, D., Pirolo, M., Lugli, G.A., Ventura, M., Frangipani, E. and Visca, P., 2020, The microbial community of a biofilm lining the wall of a pristine cave in Western New Guinea. Microbiological research, 241, 126584.
[https://doi.org/10.1016/j.micres.2020.126584]

-
Vanghi, V., Frisia, S. and Borsato, A., 2017, Genesis and microstratigraphy of calcite coralloids analysed by high resolution imaging and petrography. Sedimentary Geology, 359, 16-28.
[https://doi.org/10.1016/j.sedgeo.2017.08.001]

-
Vardeh, D.P., Woodhouse, J.N. and Neilan, B.A., 2018, Microbial diversity of speleothems in two southeast Australian limestone cave arches. Journal of Cave and Karst Studies, 80, 121-132.
[https://doi.org/10.4311/2017MB0119]

-
Vautrin, F., Pujic, P., Paquet, C., Bergeron, E., Mouniée, D., Marchal, T., Salord, H., Bonnet, J.M., Cournoyer, B., Winiarski, T., Louzier, V. and Rodriguez-Nava, V., 2021, Microbial risk assessment of Nocardia cyriacigeorgica in polluted environments, case of urban rainfall water. Computational and Structural Biotechnology Journal, 19, 384-400.
[https://doi.org/10.1016/j.csbj.2020.12.017]

-
Webster, K.D., Schimmelmann, A., Drobniak, A., Mastalerz, M., Lagarde, L.R., Boston, P.J. and Lennon, J.T., 2022, Diversity and Composition of Methanotroph Communities in Caves. Microbiology Spectrum, 10, e01566-21.
[https://doi.org/10.1128/spectrum.01566-21]

-
Wen, L., Li, D., Yang, L., Luo, P., Chen, H., Xiao, K., Song, T., Zhang, W., He, X., Chen, H. and Wang, K., 2016, Rapid recuperation of soil nitrogen following agricultural abandonment in a karst area, southwest China. Biogeochemistry, 129, 341-354.
[https://doi.org/10.1007/s10533-016-0235-3]

-
Wiseschart, A., Mhuantong, W., Tangphatsornruang, S., Chantasingh, D. and Pootanakit, K., 2019, Shotgun metagenomic sequencing from Manao-Pee cave, Thailand, reveals insight into the microbial community structure and its metabolic potential. BMC Microbiology, 19, 1-14.
[https://doi.org/10.1186/s12866-019-1521-8]

-
Yun, Y., Xiang, X., Wang, H., Man, B., Gong, L., Liu, Q., Dong, Q. and Wang, R., 2016, Five-Year Monitoring of Bacterial Communities in Dripping Water from the Heshang Cave in Central China: Implication for Paleoclimate Reconstruction and Ecological Functions. Geomicrobiology Journal, 33, 1-11.
[https://doi.org/10.1080/01490451.2015.1062062]

-
Zhang, J., Kobert, K., Flouri, T. and Stamatakis, A., 2014, PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics, 30, 614-620.
[https://doi.org/10.1093/bioinformatics/btt593]

-
Zhu, H.-Z., Jiang C.-Y. and Liu, S.-J., 2022, Microbial roles in cave biogeochemical cycling. Frontiers of Microbiology, 13, 950005.
[https://doi.org/10.3389/fmicb.2022.950005]

-
Zhu, H.-Z., Zhang, Z.-F., Zhou, N., Jiang, C.-Y., Wang, B.-J., Cai, L. and Liu, S.-J., 2019, Diversity, Distribution and Co-occurrence Patterns of Bacterial Communities in a Karst Cave System. Frontiers in Microbiology, 10, 1726.
[https://doi.org/10.3389/fmicb.2019.01726]

-
Zhu, H.-Z., Zhang, Z.-F., Zhou, N., Jiang, C.-Y., Wang, B.-J., Cai, L., Wang, H.-M. and Liu, S.-J., 2021, Bacteria and Metabolic Potential in Karst Caves Revealed by Intensive Bacterial Cultivation and Genome Assembly. Applied and Environmental Microbiology, 87, e02440-20.
[https://doi.org/10.1128/AEM.02440-20]